Nature research paper: Opposing roles of hepatic stellate cell subpopulations in hepatocarcinogenesis
Silvia AffoJorge M. Caviglia
Present address: Department of Health and Nutrition Sciences, Brooklyn College, City University of New York, New York, NY, USAPresent address: Department of Biomedical Engineering, Widener University, Chester, PA, USAThese authors contributed equally: Yoshinobu Saito, Ajay Nair, Dianne Dapito, Le-Xing YuAveline Filliol, Yoshinobu Saito, Ajay Nair, Dianne H. Dapito, Le-Xing Yu, Aashreya Ravichandra, Sonakshi Bhattacharjee, Silvia Affo, Qiuyan Sun, Jorge M.
Liver Tumor Translational Research Program, Harold C.
United States Latest News, United States Headlines
Similar News:You can also read news stories similar to this one that we have collected from other news sources.
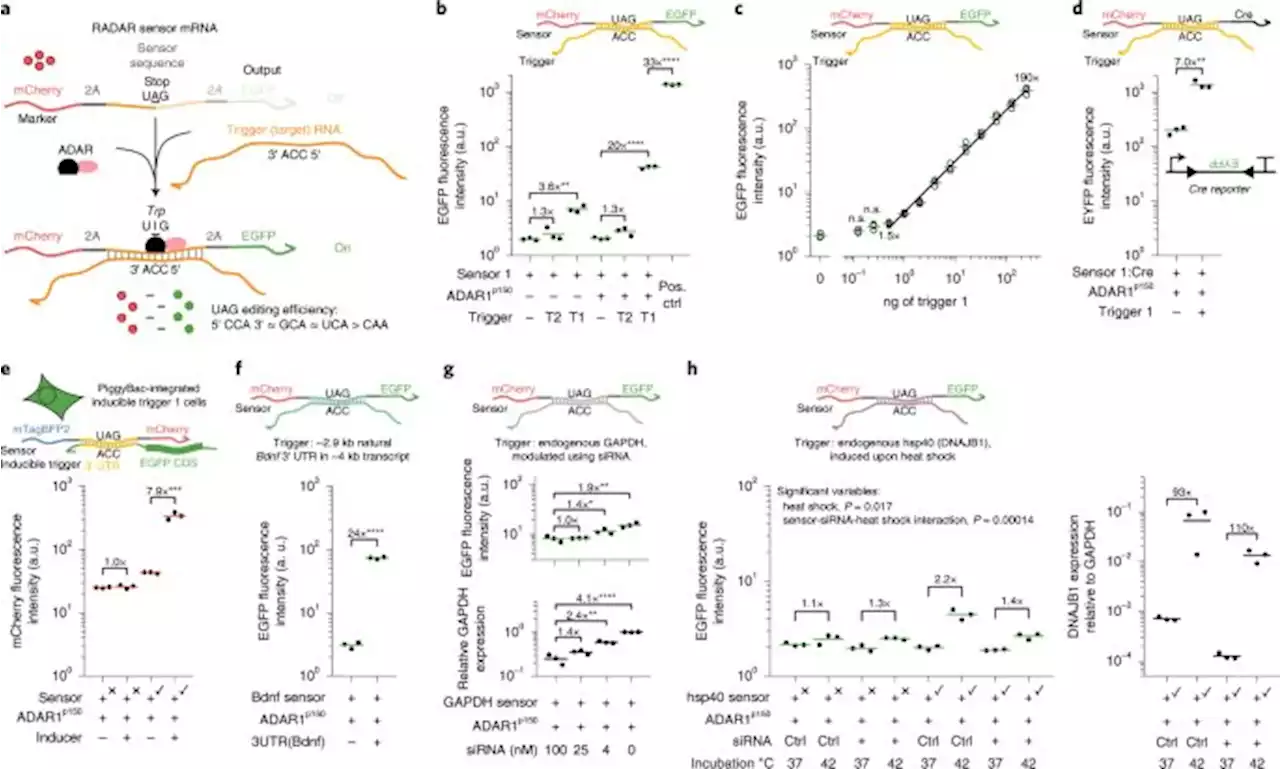 Modular, programmable RNA sensing using ADAR editing in living cells - Nature BiotechnologyModular, programmable RNA sensing using ADAR editing in living cells - SynBioGaoLab
Modular, programmable RNA sensing using ADAR editing in living cells - Nature BiotechnologyModular, programmable RNA sensing using ADAR editing in living cells - SynBioGaoLab
Read more »
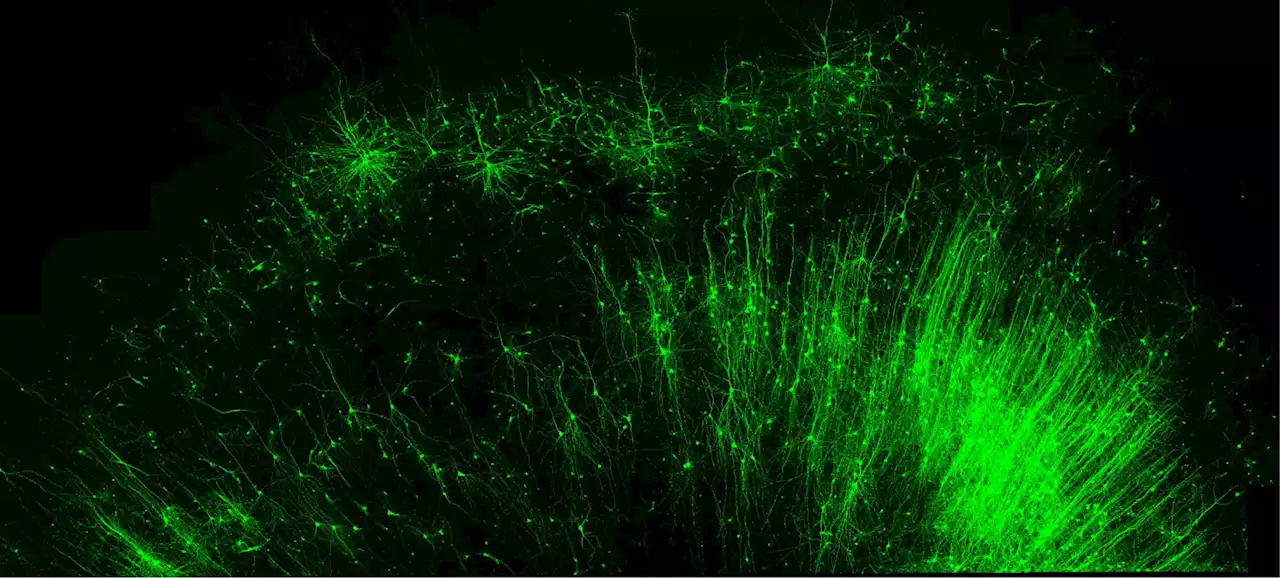 New RNA Tool Can Illuminate Brain Circuits and Edit Specific CellsEditing technology is precise and broadly applicable to all tissues and species. Scientists at Duke University have developed an RNA-based editing tool that targets individual cells, rather than genes. It is capable of precisely targeting any type of cell and selectively adding any protein of int
New RNA Tool Can Illuminate Brain Circuits and Edit Specific CellsEditing technology is precise and broadly applicable to all tissues and species. Scientists at Duke University have developed an RNA-based editing tool that targets individual cells, rather than genes. It is capable of precisely targeting any type of cell and selectively adding any protein of int
Read more »
 Biophysical Modeling of SARS-CoV-2 Assembly: Genome Condensation and BuddingThe COVID-19 pandemic caused by the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) has spurred unprecedented and concerted worldwide research to curtail and eradicate this pathogen. SARS-CoV-2 has four structural proteins: Envelope (E), Membrane (M), Nucleocapsid (N), and Spike (S), which self-assemble along with its RNA into the infectious virus by budding from intracellular lipid membranes. In this paper, we develop a model to explore the mechanisms of RNA condensation by structural proteins, protein oligomerization and cellular membrane–protein interactions that control the budding process and the ultimate virus structure. Using molecular dynamics simulations, we have deciphered how the positively charged N proteins interact and condense the very long genomic RNA resulting in its packaging by a lipid envelope decorated with structural proteins inside a host cell. Furthermore, considering the length of RNA and the size of the virus, we find that the intrinsic curvature of M proteins is essential for virus budding. While most current research has focused on the S protein, which is responsible for viral entry, and it has been motivated by the need to develop efficacious vaccines, the development of resistance through mutations in this crucial protein makes it essential to elucidate the details of the viral life cycle to identify other drug targets for future therapy. Our simulations will provide insight into the viral life cycle through the assembly of viral particles de novo and potentially identify therapeutic targets for future drug development.
Biophysical Modeling of SARS-CoV-2 Assembly: Genome Condensation and BuddingThe COVID-19 pandemic caused by the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) has spurred unprecedented and concerted worldwide research to curtail and eradicate this pathogen. SARS-CoV-2 has four structural proteins: Envelope (E), Membrane (M), Nucleocapsid (N), and Spike (S), which self-assemble along with its RNA into the infectious virus by budding from intracellular lipid membranes. In this paper, we develop a model to explore the mechanisms of RNA condensation by structural proteins, protein oligomerization and cellular membrane–protein interactions that control the budding process and the ultimate virus structure. Using molecular dynamics simulations, we have deciphered how the positively charged N proteins interact and condense the very long genomic RNA resulting in its packaging by a lipid envelope decorated with structural proteins inside a host cell. Furthermore, considering the length of RNA and the size of the virus, we find that the intrinsic curvature of M proteins is essential for virus budding. While most current research has focused on the S protein, which is responsible for viral entry, and it has been motivated by the need to develop efficacious vaccines, the development of resistance through mutations in this crucial protein makes it essential to elucidate the details of the viral life cycle to identify other drug targets for future therapy. Our simulations will provide insight into the viral life cycle through the assembly of viral particles de novo and potentially identify therapeutic targets for future drug development.
Read more »
 Liver Fat Lowered by Ezetimibe-Statin CombinationA significant 5.8% decrease in hepatic steatosis was seen after 24 weeks’ treatment with ezetimibe and rosuvastatin comparing baseline with end of treatment values.
Liver Fat Lowered by Ezetimibe-Statin CombinationA significant 5.8% decrease in hepatic steatosis was seen after 24 weeks’ treatment with ezetimibe and rosuvastatin comparing baseline with end of treatment values.
Read more »
 Inferring and perturbing cell fate regulomes in human brain organoids - NatureA multi-omic atlas of brain organoid development facilitates the inference of an underlying gene regulatory network using the newly developed Pando framework and shows—in conjunction with perturbation experiments—that GLI3 controls forebrain fate establishment through interaction with HES4/5 regulomes.
Inferring and perturbing cell fate regulomes in human brain organoids - NatureA multi-omic atlas of brain organoid development facilitates the inference of an underlying gene regulatory network using the newly developed Pando framework and shows—in conjunction with perturbation experiments—that GLI3 controls forebrain fate establishment through interaction with HES4/5 regulomes.
Read more »
 Modular, programmable RNA sensing using ADAR editing in living cells - Nature BiotechnologyModular, programmable RNA sensing using ADAR editing in living cells - SynBioGaoLab
Modular, programmable RNA sensing using ADAR editing in living cells - Nature BiotechnologyModular, programmable RNA sensing using ADAR editing in living cells - SynBioGaoLab
Read more »
