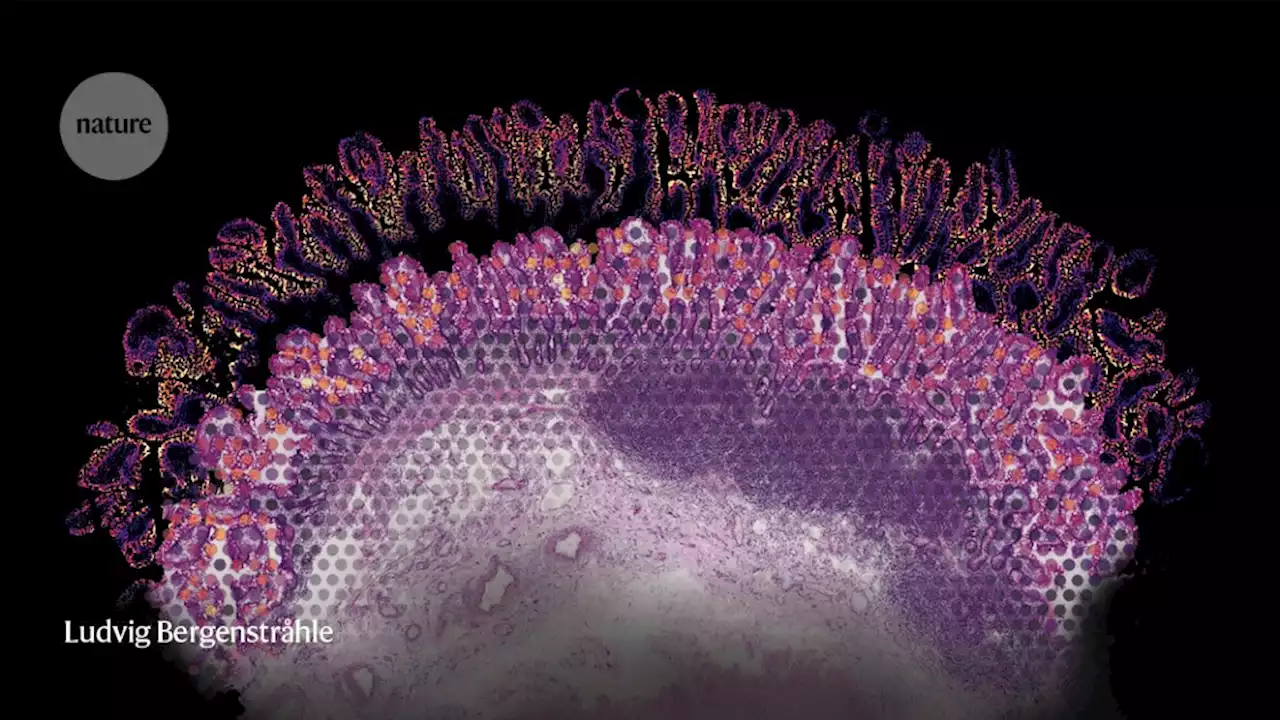
Computational and experimental methods are bringing researchers closer to their goal of revealing exactly where in a cell or tissue each gene is expressed.
Under a microscope, mammalian tissues reveal their intricate and elegant architectures. But if you look at the same tissue after tumour formation, you will see bedlam. Itai Yanai, a computational biologist at New York University’s Grossman School of Medicine in New York City, is trying to find order in this chaos. “There is a particular logic to how things are arranged, and spatial transcriptomics is helping us see that,” he says.
By contrast, spatial transcriptomics allows researchers to study gene expression in intact samples, opening frontiers in cancer research and revealing previously inaccessible biology of otherwise well-characterized tissues. The resulting ‘atlases’ of spatial information can tell scientists which cells make up each tissue, how they are organized and how they communicate.
By 1998, thanks to advances in microscopy and image processing, researchers could identify individual RNA molecules in cells. Using this single-molecule fluorescencehybridization method, it was possible to visualize individual mRNA transcripts from several genes simultaneously by using probes of different colours. But early versions of smFISH could monitor only three or four genes at a time — far short of the tens of thousands of genes expressed in the human transcriptome.
When a read-out probe binds to the read-out sequence of an encoding probe and gives off a fluorescent signal, it is read as a ‘1’; if there is no fluorescence, it is read as a ‘0’. Multiple rounds of imaging yield a binary barcode that can identify the detected RNA. The ‘error-robust’ part of the technique refers to the barcodes’ design: they are sufficiently different from one other that there is little chance of misinterpreting which mRNA sequence is being detected.
United States Latest News, United States Headlines
Similar News:You can also read news stories similar to this one that we have collected from other news sources.
 Nura’s New Earphones Offer CD-Quality Lossless Audio And Adaptive Noise CancellationNura’s new earphones offer personalized sound, CD-quality lossless audio, spatial processing and adaptive noise cancellation.
Nura’s New Earphones Offer CD-Quality Lossless Audio And Adaptive Noise CancellationNura’s new earphones offer personalized sound, CD-quality lossless audio, spatial processing and adaptive noise cancellation.
Read more »
 What Is CRISPR, and Why Is It So Important?It's been 10 years since the debut of CRISPR. The revolutionary gene editing tool has taken off, but scientists are still far from realizing its potential
What Is CRISPR, and Why Is It So Important?It's been 10 years since the debut of CRISPR. The revolutionary gene editing tool has taken off, but scientists are still far from realizing its potential
Read more »
 BMW To Start Using Android Automotive OS Next Year | CarscoopsBMW To Start Using Android Automotive OS Next Year | Carscoops carscoops
BMW To Start Using Android Automotive OS Next Year | CarscoopsBMW To Start Using Android Automotive OS Next Year | Carscoops carscoops
Read more »
 Supreme Court Blocks Order Creating Second Majority-Black District in LouisianaThe Supreme Court granted an application from Republican Louisiana officials to block a district court order that the state create a second majority-Black congressional district
Supreme Court Blocks Order Creating Second Majority-Black District in LouisianaThe Supreme Court granted an application from Republican Louisiana officials to block a district court order that the state create a second majority-Black congressional district
Read more »
 Over 4K without power in Houston areaAccording to the CenterPoint Energy outage map, 10,531 people are without power, as of 8:35 p.m.
Over 4K without power in Houston areaAccording to the CenterPoint Energy outage map, 10,531 people are without power, as of 8:35 p.m.
Read more »