An article published in GenomeBiology discusses EvoAug: a suite of evolution-inspired augmentations that enhance the training of genomic Deep neural networks by increasing genetic variation.
EvoAug is comprised of a set of data augmentations given by the following:
Mutation: a transformation where single nucleotide mutations are randomly applied to a given wild-type sequence. This is implemented as follows: given the hyperparameter of the fraction of nucleotides in each sequence to mutate , the number of mutations for a given sequence length is calculated; a position along the sequence is randomly sampled for each number of mutations; and the selected positions are mutagenized to a random nucleotide.
Deletion: a transformation where a random, contiguous segment of a wild-type sequence is removed, and the shortened sequence is then padded with random DNA sequence to maintain the same length as wild-type.
Inversion: a transformation where a random subsequence is replaced by its reverse-complement.
For the Basset and DeepSTARR models, each augmentation has an optimal setting that was determined from a hyperparameter search independently using the validation set . For the Basset models, the hyperparameters were set to:
United States Latest News, United States Headlines
Similar News:You can also read news stories similar to this one that we have collected from other news sources.
 Location and condition based reconstruction of colon cancer microbiome from human RNA sequencing data - Genome MedicineBackground The association between microbes and cancer has been reported repeatedly; however, it is not clear if molecular tumour properties are connected to specific microbial colonisation patterns. This is due mainly to the current technical and analytical strategy limitations to characterise tumour-associated bacteria. Methods Here, we propose an approach to detect bacterial signals in human RNA sequencing data and associate them with the clinical and molecular properties of the tumours. The method was tested on public datasets from The Cancer Genome Atlas, and its accuracy was assessed on a new cohort of colorectal cancer patients. Results Our analysis shows that intratumoural microbiome composition is correlated with survival, anatomic location, microsatellite instability, consensus molecular subtype and immune cell infiltration in colon tumours. In particular, we find Faecalibacterium prausnitzii, Coprococcus comes, Bacteroides spp., Fusobacterium spp. and Clostridium spp. to be strongly associated with tumour properties. Conclusions We implemented an approach to concurrently analyse clinical and molecular properties of the tumour as well as the composition of the associated microbiome. Our results may improve patient stratification and pave the path for mechanistic studies on microbiota-tumour crosstalk.
Location and condition based reconstruction of colon cancer microbiome from human RNA sequencing data - Genome MedicineBackground The association between microbes and cancer has been reported repeatedly; however, it is not clear if molecular tumour properties are connected to specific microbial colonisation patterns. This is due mainly to the current technical and analytical strategy limitations to characterise tumour-associated bacteria. Methods Here, we propose an approach to detect bacterial signals in human RNA sequencing data and associate them with the clinical and molecular properties of the tumours. The method was tested on public datasets from The Cancer Genome Atlas, and its accuracy was assessed on a new cohort of colorectal cancer patients. Results Our analysis shows that intratumoural microbiome composition is correlated with survival, anatomic location, microsatellite instability, consensus molecular subtype and immune cell infiltration in colon tumours. In particular, we find Faecalibacterium prausnitzii, Coprococcus comes, Bacteroides spp., Fusobacterium spp. and Clostridium spp. to be strongly associated with tumour properties. Conclusions We implemented an approach to concurrently analyse clinical and molecular properties of the tumour as well as the composition of the associated microbiome. Our results may improve patient stratification and pave the path for mechanistic studies on microbiota-tumour crosstalk.
Read more »
 Protein fibril length in cerebrospinal fluid is increased in Alzheimer’s disease - Communications BiologyLiquid-based AFM on protein aggregates at single-fibril resolution from the cerebrospinal fluid of patients at various stages of AD reveals that mature fibrils from patients at advanced stage AD have longer length, distinct morphology and spatial organization.
Protein fibril length in cerebrospinal fluid is increased in Alzheimer’s disease - Communications BiologyLiquid-based AFM on protein aggregates at single-fibril resolution from the cerebrospinal fluid of patients at various stages of AD reveals that mature fibrils from patients at advanced stage AD have longer length, distinct morphology and spatial organization.
Read more »
 More diverse gene map could lead to better treatmentsResearchers produce a new version of the human genome that could improve medical treatments.
More diverse gene map could lead to better treatmentsResearchers produce a new version of the human genome that could improve medical treatments.
Read more »
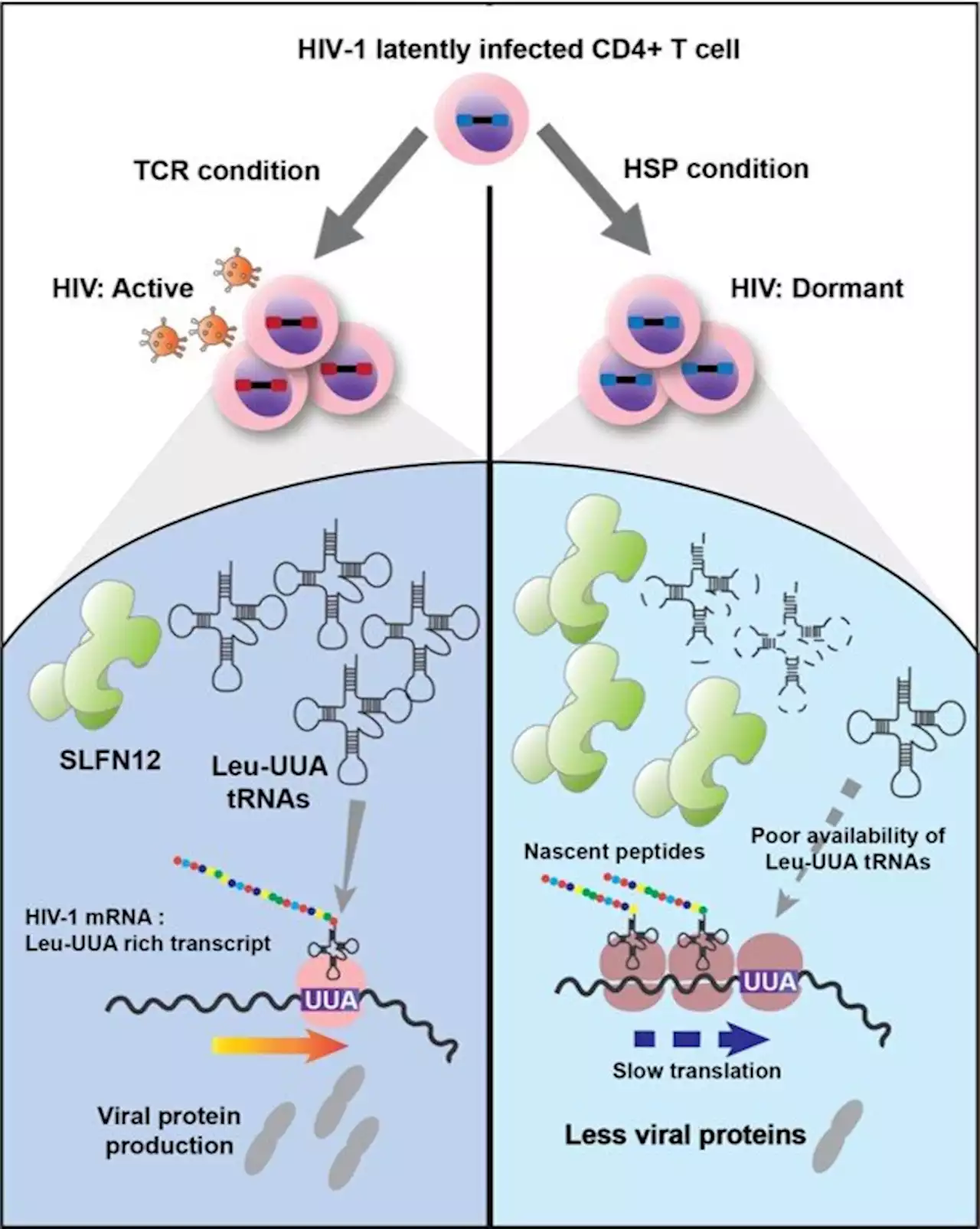 One step closer to eliminating latency, the real challenge in combating HIVAn international study led by MELIS-UPF researchers from the Infection Biology and Molecular Virology laboratories has identified and characterized Schlafen 12 (SLFN 12) as a novel HIV restriction factor. SLFN 12 shuts down viral protein production and helps virus-infected cells to escape from anti-HIV therapy and immune responses. These findings pave the way for improving therapeutic strategies that aim to cure HIV infections.
One step closer to eliminating latency, the real challenge in combating HIVAn international study led by MELIS-UPF researchers from the Infection Biology and Molecular Virology laboratories has identified and characterized Schlafen 12 (SLFN 12) as a novel HIV restriction factor. SLFN 12 shuts down viral protein production and helps virus-infected cells to escape from anti-HIV therapy and immune responses. These findings pave the way for improving therapeutic strategies that aim to cure HIV infections.
Read more »
 High-grade gliomas remodel neural circuits: promoting tumor progression and impaired cognitionHigh-grade gliomas remodel neural circuits: promoting tumor progression and impaired cognition Nature UCSF gliomas glioblastoma tumor cognition neuroscience
High-grade gliomas remodel neural circuits: promoting tumor progression and impaired cognitionHigh-grade gliomas remodel neural circuits: promoting tumor progression and impaired cognition Nature UCSF gliomas glioblastoma tumor cognition neuroscience
Read more »
 Location and condition based reconstruction of colon cancer microbiome from human RNA sequencing data - Genome MedicineBackground The association between microbes and cancer has been reported repeatedly; however, it is not clear if molecular tumour properties are connected to specific microbial colonisation patterns. This is due mainly to the current technical and analytical strategy limitations to characterise tumour-associated bacteria. Methods Here, we propose an approach to detect bacterial signals in human RNA sequencing data and associate them with the clinical and molecular properties of the tumours. The method was tested on public datasets from The Cancer Genome Atlas, and its accuracy was assessed on a new cohort of colorectal cancer patients. Results Our analysis shows that intratumoural microbiome composition is correlated with survival, anatomic location, microsatellite instability, consensus molecular subtype and immune cell infiltration in colon tumours. In particular, we find Faecalibacterium prausnitzii, Coprococcus comes, Bacteroides spp., Fusobacterium spp. and Clostridium spp. to be strongly associated with tumour properties. Conclusions We implemented an approach to concurrently analyse clinical and molecular properties of the tumour as well as the composition of the associated microbiome. Our results may improve patient stratification and pave the path for mechanistic studies on microbiota-tumour crosstalk.
Location and condition based reconstruction of colon cancer microbiome from human RNA sequencing data - Genome MedicineBackground The association between microbes and cancer has been reported repeatedly; however, it is not clear if molecular tumour properties are connected to specific microbial colonisation patterns. This is due mainly to the current technical and analytical strategy limitations to characterise tumour-associated bacteria. Methods Here, we propose an approach to detect bacterial signals in human RNA sequencing data and associate them with the clinical and molecular properties of the tumours. The method was tested on public datasets from The Cancer Genome Atlas, and its accuracy was assessed on a new cohort of colorectal cancer patients. Results Our analysis shows that intratumoural microbiome composition is correlated with survival, anatomic location, microsatellite instability, consensus molecular subtype and immune cell infiltration in colon tumours. In particular, we find Faecalibacterium prausnitzii, Coprococcus comes, Bacteroides spp., Fusobacterium spp. and Clostridium spp. to be strongly associated with tumour properties. Conclusions We implemented an approach to concurrently analyse clinical and molecular properties of the tumour as well as the composition of the associated microbiome. Our results may improve patient stratification and pave the path for mechanistic studies on microbiota-tumour crosstalk.
Read more »
